Bioenergetics and Quinones
We study the origins and diversification of metabolic pathways involved in bioenergetics using phylogenomics and experimental approaches.
We investigate in particular, the evolution of the different biosynthetic pathways of quinones (find our recent review here). Quinones are molecules involved in electron transfer chains. They are therefore crucial for cellular energy production.
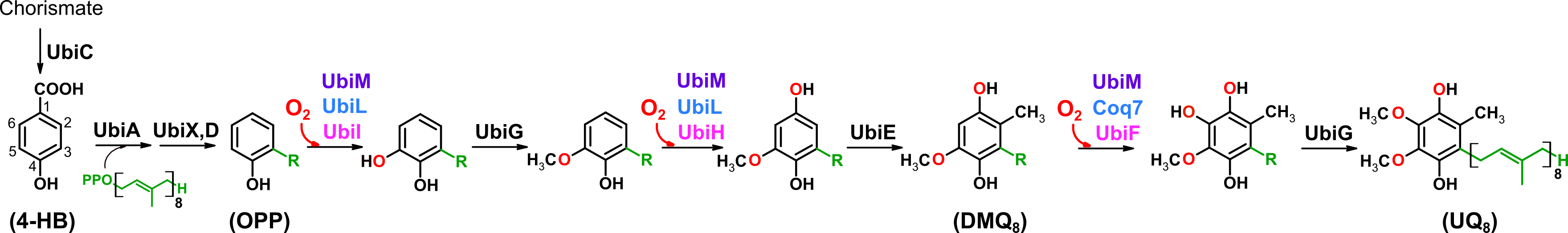
Variability of the ubiquinone biosynthetic pathway. In particular, three hydroxylation steps can be performed by different sets of enzymes depending on the species. In purple, UbiM performs the three hydroxylation steps (as in Neisseria meningitidis). In blue, two enzymes are required, as in Rhodobacter capsulatus. In red, three enzymes are required, as in Escherichia coli. Figure from Kazemzadeh et al. Mol. Biol. Evol, 2023.
We also currently study the evolution of a particular energy metabolism: phototrophy.
We combine experimental and computational approaches. This goes from developing tools for genome annotation of the proteins involved in the pathways, to the study of their functional diversification, and in vivo testing of their predicted functions.
The QuinEvol project
An example of study that combines experimental and computational approaches was recently published. We show the diversity of the pathway used by Pseudomonadota (ex-proteobacteria) to produce ubiquinone, and at the same time explore the diversification of the flavin monooxygenase (FMO) protein family by gene transfers and duplications.
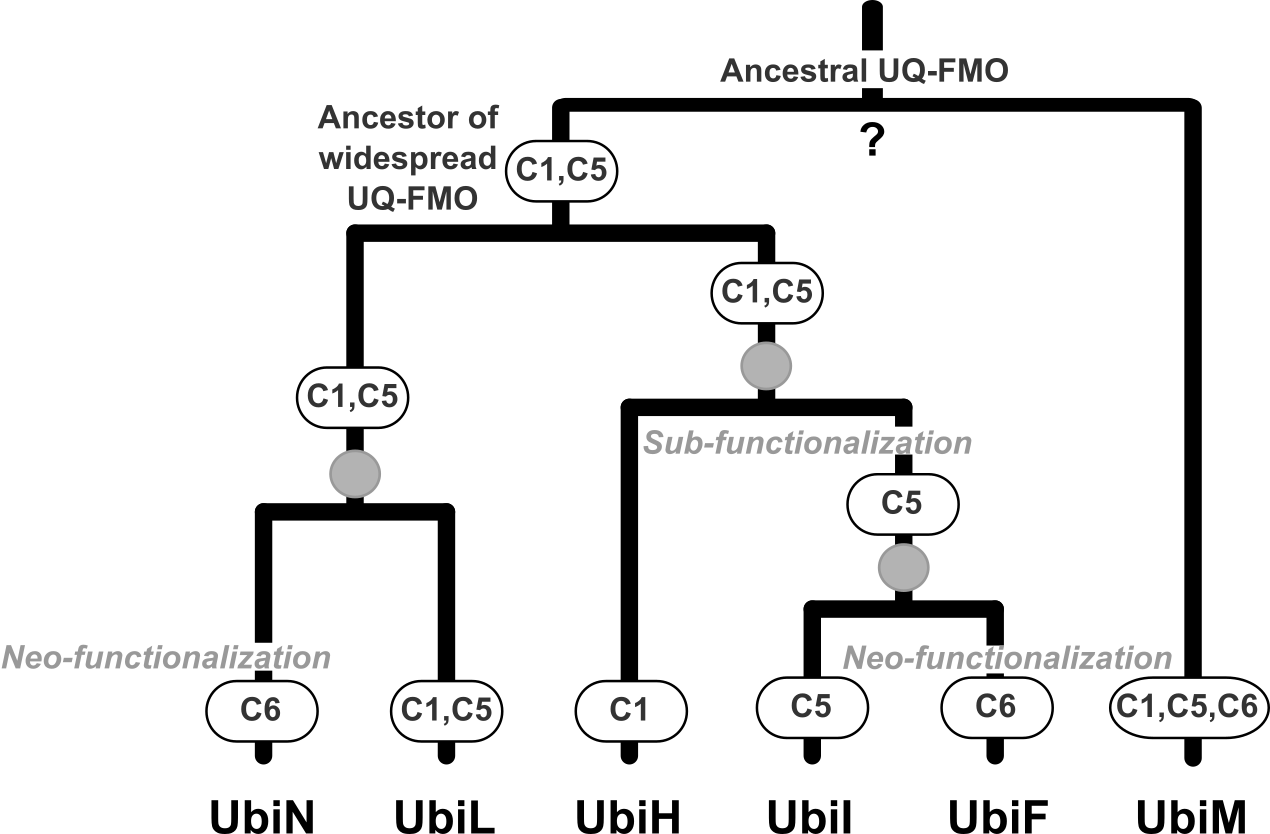
Evolution and diversification of the UQ-FMO protein family. Members of this family of hydroxylases evolved by duplication different regio-selectivities within the ubiquinone biosynthetic pathway - being able to hydroxylase Carbon 1, 5 and/or 6 of the UQ precursor. Figure from Kazemzadeh et al. Mol. Biol. Evol, 2023.
Team members also recently characterized a new pathway for ubiquinone production, which is O2-independent (contrary to the classical, O2-dependent one). We could recently contribute to understanding its origin and physiological role.
We recently published a study on the repertoire of quinones found in Pseudomonadota and how the dynamics of this repertoire likely accompagnied the diversification of aerobic, facultative aerobic, microaerophilic and anaerobic metabolisms. To this end, we proposed an evolutionary scenario to explain the variable content in quinones across Pseudomonadota. 
Global evolutionary scenario for the quinone repertoire of Pseudomonadota. For full legend see here.
Finally, we could show that Cyanobacteria inherited from Pseudomonadota an enzyme that is crucial for the production of the plastoquinone (PQ), used in photosynthetic chains. This enzyme is a flavin monooxygenase, and we show it requires O2 to proceed. Hence, most contemporary Cyanobacteria need O2 for oxygenic photosynthesis. This work was recently published at the FEBS Journal.
The QuinEvol project was funded by the ANR (2021-2025).
The AbiSYM project
Now that we understand well the evolution of the ubiquinone biosynthetic pathway in Pseudomonadota, we are currently investigating how comes that eukaryotes did not inherit in their mitochondria, the exact same UQ pathway that is found in the lineages that are close to the ancestor of the mitochondrion. Answering this question might bring light on the establishment of bioenergetic processes during eukaryogenesis, and in particular, the potential contribution of other prokaryotic contributors than Pseudomonadota.
The AbiSYM project is funded by the ANR (2024-2029).
The ToLQuin project
We recently contributed to a study that reports the discovery of a novel quinone, the methyl-plastoquinone, in the phylum Nitrospirota. This was published in PNAS in 2025.
In a newly funded project, together with our partner Dr. Felix Elling (U. Kiel, Germany), we will investigate the possibility of novel quinones in the tree of life, explore lowly characterized quinone pathways, and investigate the impact of quinone evolution on the diversification of energy metabolism.
The ToLQuin project is funded by the ANR and the DFG (2026-2029). It will officially start in May 2026. A post-doc position will soon be announced.
The ADAPT2Q project
In collaboration with the BIP and the LCB labs in Marseille, we are currently working on the ADAPT2Q ANR-funded project that aims at understanding the adaptation of quinone-interacting enzymes to the diversity of quinones that have evolved along time.
The ADAPT2Q project is funded by the ANR (2023-2027).
Funders
These projects are mainly funded by the French National Research Agency, “ANR”.

