Genome Annotation
We develop software and methods to improve genome annotations.
MacSyFinder, the macromolecular system finder
In particular we developed MacSyFinder, a software to:
- model molecular systems (pathways, machineries…) in microbial genomes
- annotate them in genomes
MacSyFinder takes advantage of the genomic organization by function of archaeal and bacterial genomes: genes participating in a same function, complex, pathway… tend to co-localize on the genome. This enables powerful, system-level (instead of gene-by-gene) annotation of functions.
For more details, have a look at the corresponding publication(s).
The MacSyFinder v2 search engine
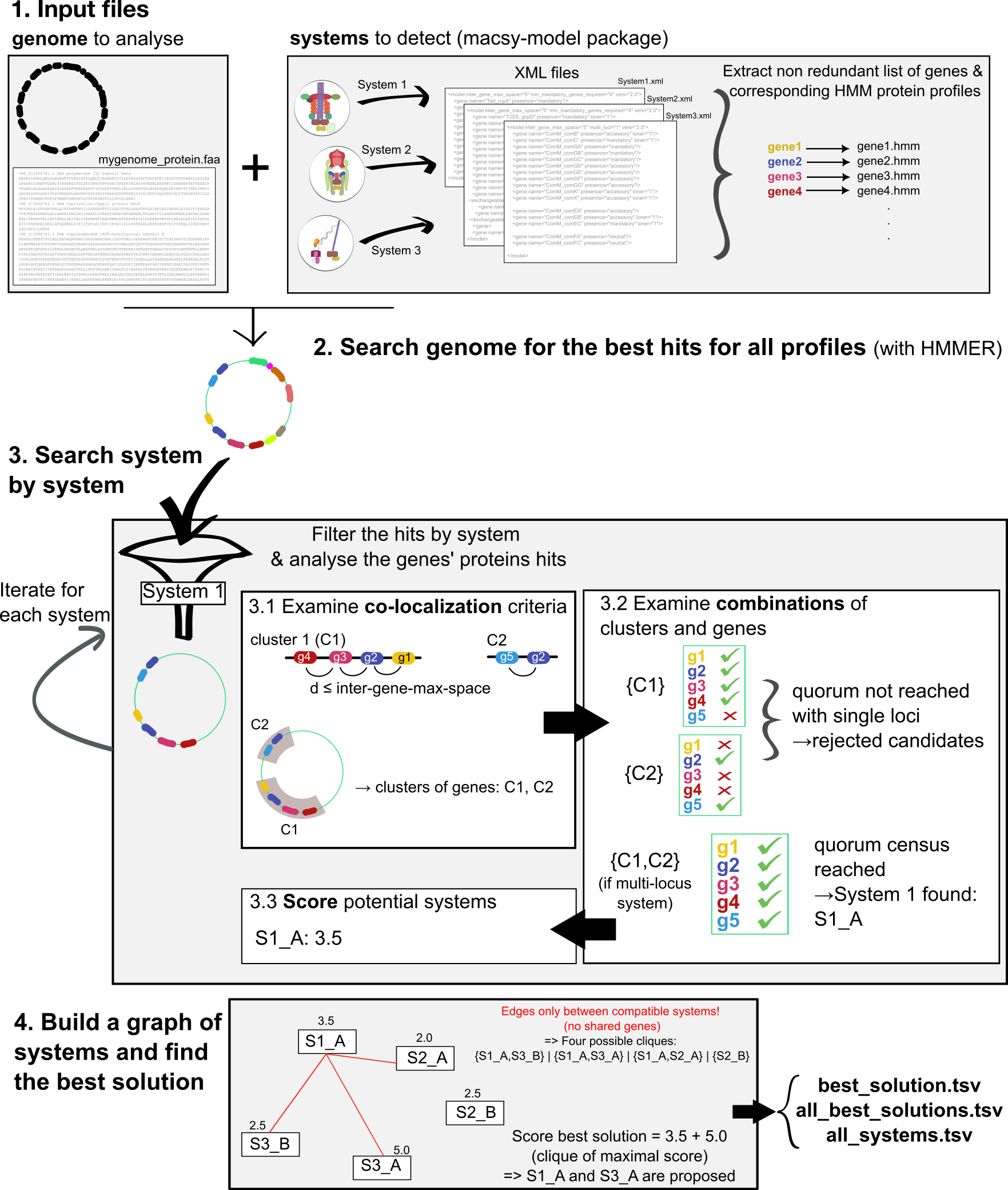
Figure from Néron et. al 2023., the MSF v2 paper reviewed and recommended by PCI Genomics.
MacsyModels, an organization for sharing MacSyFinder models
Since MacSyFinder version 2, it is possible to easily download, install and share publicly available MacSyFinder models. This is done from the official MacSy-models organization. Several models are readily available, as described in Néron et. al 2023..
As of March 2023, Macsy Models included models for the detection of:
- CRISPR-Cas systems CasFinder
- bacterial protein secretion systems and type IV-filament super-family members TXSScan
- conjugative systems CONJScan
For more on how to install MacSyFinder and the associated Macsy Models, have a look at the comprehensive MacSyFinder’s documentation.
Writing own MacsyModels
In the following publication, we explain step-by-step how one can write its own MacsyModels (models’ definition and profiles). If interested, have a look here:
Abby, Denise and Rocha 2024 Identification of Protein Secretion Systems in Bacterial Genomes Using MacSyFinder Version 2, 2024 https://link.springer.com/10.1007/978-1-0716-3445-5_1
Machine learning for genome annotation
Together with Nelle Varoquaux, we currently develop machine learning approaches for genome annotation. We supervise a PhD student in the frame of this project that is supported by the ANR program agency. More soon!

